Detecting beats in the photoplethysmogram: benchmarking open-source algorithms
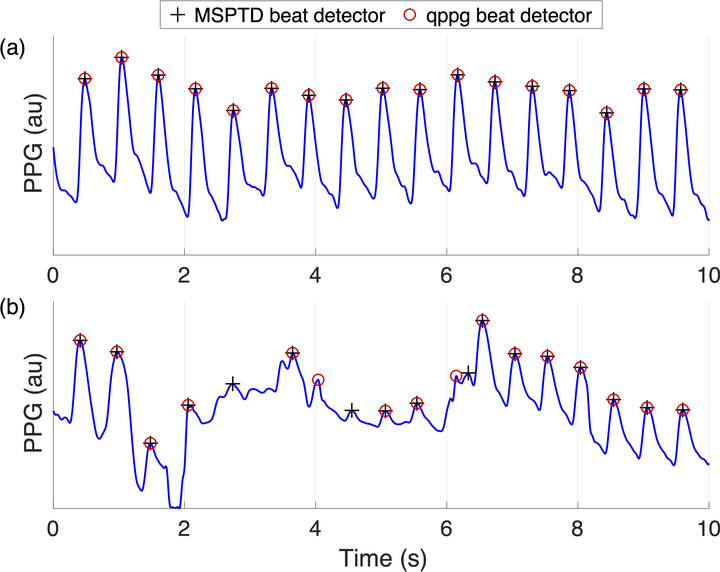 Image credit: P.H. Charlton et al. (CC BY 4.0)
Image credit: P.H. Charlton et al. (CC BY 4.0)
Abstract
The photoplethysmogram (PPG) signal is widely used in pulse oximeters and smartwatches. A fundamental step in analysing the PPG is the detection of heartbeats. Several PPG beat detection algorithms have been proposed, although it is not clear which performs best. Objective: This study aimed to: (i) develop a framework with which to design and test PPG beat detectors; (ii) assess the performance of PPG beat detectors in different use cases; and (iii) investigate how their performance is affected by patient demographics and physiology. Approach: Fifteen beat detectors were assessed against electrocardiogram-derived heartbeats using data from eight datasets. Performance was assessed using the F1 score, which combines sensitivity and positive predictive value. Main results: Eight beat detectors performed well in the absence of movement with F1 scores of ≥90% on hospital data and wearable data collected at rest. Their performance was poorer during exercise with F1 scores of 55-91%; poorer in neonates than adults with F1 scores of 84-96% in neonates compared to 98-99% in adults; and poorer in atrial fibrillation (AF) with F1 scores of 92-97% in AF compared to 99-100% in normal sinus rhythm. Significance: Two PPG beat detectors denoted ‘MSPTD’ and ‘qppg’ performed best, with complementary performance characteristics. This evidence can be used to inform the choice of PPG beat detector algorithm. The algorithms, datasets, and assessment framework are freely available.
Accompanying Resources
This study used two key resources:
- The MIMIC PERform datasets.
- The ‘ppg-beats’ toolbox of PPG beat detection algorithms and code to assess their performance.
Reproducing the analysis
The analysis reported in this study can be reproduced by following these steps:
Download the required datasets
The datasets used in the study are listed on this page. Download each dataset in turn by following the instructions provided on each dataset’s page.
Install the PPG-beats toolbox
Install the PPG-beats toolbox by following the instructions on this page.
Assess the performance of PPG beat detectors across all the datasets
Use the assess_multiple_datasets.m script (within PPG-beats) to assess the performance of PPG beat detectors across all of the datasets. To do so:
- Edit the
assess_multiple_datasets.mscript:- Specify the path of the Matlab file for each dataset in the
specify_path_of_dataset_filefunction within theassess_multiple_datasets.mscript. - Specify the folder in which to save results figures by modifying the
up.paths.plots_root_foldervariable within thesetup_universal_paramsfunction within theassess_multiple_datasets.mscript. - Specify the datasets to be analysed in the
setup_universal_paramsfunction within theassess_multiple_datasets.mscript:
up.assessment_datasets = {'capnobase', 'bidmc', 'mimic_train_all', 'mimic_test_all', 'wesad_meditation', 'wesad_amusement', 'wesad_baseline', 'wesad_stress', 'ppg_dalia_sitting', 'ppg_dalia_working', 'ppg_dalia_cycling', 'ppg_dalia_walking', 'ppg_dalia_lunch_break', 'ppg_dalia_car_driving', 'ppg_dalia_stair_climbing', 'ppg_dalia_table_soccer'}; up.comparison_datasets = {'mimic_B', 'mimic_W', 'mimic_test_a', 'mimic_test_n', 'mimic_non_af', 'mimic_af'};- Specify the beat detectors to be used in the
specify_optionsfunction within theassess_multiple_datasets.mscript:
options.beat_detectors = {'SWT', 'ATmax', 'SPAR', 'IMS', 'AMPD', 'MSPTD', 'ABD', 'qppgfast', 'HeartPy', 'COppg', 'PPGPulses', 'ERMA', 'PWD', 'PDA', 'WFD'}; - Specify the path of the Matlab file for each dataset in the
- Run the
assess_multiple_datasets.mscript, which will:- Analyse each dataset in turn
- Generate results (in the command window) and results figures (saved in the specified folder)
Generate additional figures
The additional figures in the manuscript can be generated as follows:
- Fig. 1 (Detecting beats in the photoplethysmogram): - run the
create_ppg_beat_detection_fig.mscript. - Fig. 2 (Comparing PPG-derived beats with reference beats): - run the
make_plot_of_ecg_ppg_time_alignment.mscript. - Fig. 5 (PPG beat detection during different activities): - run the
make_plot_of_ppg_beat_detection_challenges.mscript.